RAG + Langchain – Interacting with PDF documents using prompts
Experimenting with OpenAI LLM to extract info from PDFs through prompts
I have been researching the topic of RAGs lately so I thought of tinkering on a project. As I was working on my Master’s Thesis, I found the task of adding citations quite cumbersome. I would read several papers as I worked through the code and report, but when it came to citations, I had to manually find the papers, get the author’s names and titles and format them in the required standard.
So I wondered whether it was possible to automate this task - i.e. given a set of PDFs, extract the relevant info and format them in a specific citation standard.
Tech and tools used for this project
- RAG
- PyPDFLoader
- Vector Database (ChromaDB), and SQLite3
- Langchain
- OpenAI
- Streamlit
- Docker
Quick note on RAG
RAG stands for Retrieval Augmented Generation. It is a way of retrieving specific, relevant information from from a database or a set of documents (like PDFs or text files) and using natural language processing (NLP) and large language models (LLMs) to generate an answer to a user query.
If you prompt ChatGPT with a question, it can respond based on its training, which may not include your specific document, or it may hallucinate. Using RAGs allows LLMs to provide more accurate and contextually relevant responses based on real, user-provided data.
The structure of this project
1. Load and process PDF
First, take the PDF and extract the content using PyPDFLoader.
loader = PyPDFLoader(file.name)
documents = loader.load()
2. Split document into chunks
The documents on their whole can be huge, and need to be split into smaller chunks so that the LLM model can efficiently search for the most relevant parts to retrieve and base their responses on. This is done using the RecursiveCharacterTextSplitter function in the langchain framework.
text_splitter = RecursiveCharacterTextSplitter(chunk_size=chunk_size,
chunk_overlap=chunk_overlap,
length_function=len,
separators=["\n\n", "\n", " "])
chunk_size is the length of each chunk, chunk_overlap is the text that is common between each neighbouring chunk and separators indicate where the chunks should be separated (we don’t want a chunk to end and another to start mid-sentence).
3. Embedding the data
The text in each chunk has to be converted to vector embeddings – numeric representations of text in a multi-dimensional space. Think of how numerical representations of the words “Berlin” and “currywurst” may be closer in distance to each other than “Berlin” and “pasta” for example, or how those of “Paris” and “cafe” may be closer than “Paris” and “fishing”.
This embedding is to help the LLM model calculate how similar the different chunks are to the query provided (which will also be converted into a vector embedding later) so that the most relevant chunks from the document can be used for the final response.
There are various embedding models that can be used here and they vary in their accuracy and effectiveness. For this project, I have the text-embedding-ada-002 model from OpenAIEmbeddings
embeddings = OpenAIEmbeddings(
model="text-embedding-ada-002", openai_api_key=api_key
)
4. Storing the embeddings in a vector database
The vector representations need to be stored in a vector database, which is a special kind of database for embeddings. This is the database from where the chunks most relevant to a query are retrieved to generate the response.
For this project, I have used the ChromaDB database. FAISS is another option that you can try.
vectorstore = Chroma.from_documents(documents=unique_chunks,
collection_name=clean_filename(file_name),
embedding=embedding_function,
ids=list(unique_ids),
persist_directory = vector_store_path)
ids help keep track of each chunk in a document, and ensure that only unique documents with unique ids are stored. This is to avoid duplication – when 2 documents may end up having the same id.
5. Querying and generating responses
Ideally what I want next is for the user to enter a query and for the most relevant chunks from the vector store database to be retrieved using a similarity search. From these relevant chunks, a language model should then generates an answer. For this project, I chose the LLM to be the gpt-4o-mini model from OpenAI. And since I wanted to just extract basic metadata such as the titles and authors from the papers, I preset the query as “Give me the title, summary, publication date, and authors of the research paper.” and the user only has to click on a button “Generate table” to retrieve this data in a tabular format.
llm = ChatOpenAI(model="gpt-4o-mini", api_key=api_key)
retriever=vectorstore.as_retriever(search_type="similarity")
prompt_template = ChatPromptTemplate.from_template(PROMPT_TEMPLATE)
rag_chain = (
{"context": retriever | format_docs, "question": RunnablePassthrough()}
| prompt_template
| llm.with_structured_output(ExtractedInfoWithSources, strict=True)
)
structured_response = rag_chain.invoke(query)
The rag_chain command automatically chains together the steps.
6. Building an interactive frontend with Streamlit
To make the process interactive for the user, I created a simple dashboard on Streamlit with an upload option, where the user can enter their OpenAI API key, upload a PDF file (which can also be displayed as an iFrame element), and the button to generate the table with the data.
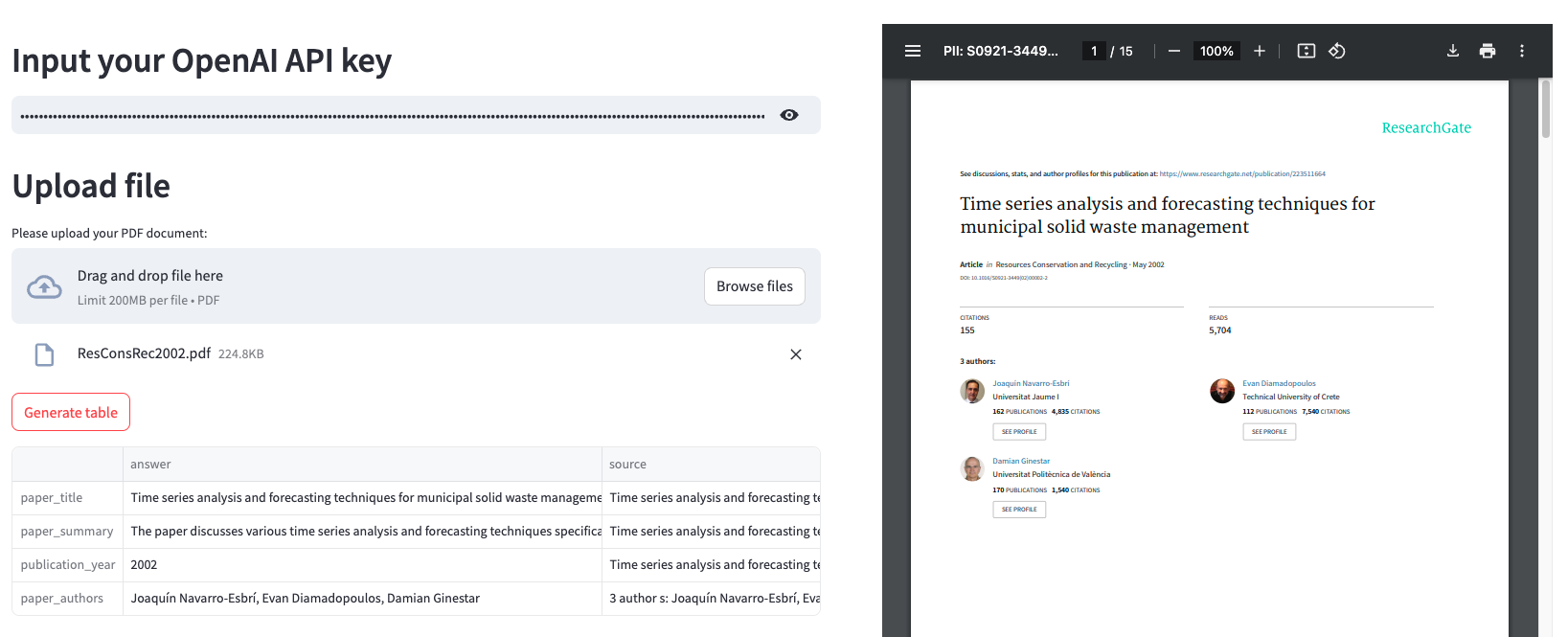
7. Deployment with Docker
Initially I thought of deploying on Streamlit Cloud but I was getting some errors pertaining to incompatibility in some packages. This was most likely ChromaDB, which requires certain specific versions of sqlite3, but the version on Streamlit Cloud seems to be older. Unfortunately, in serverless environments like Streamlit Cloud, you cannot typically control how system-level packages like sqlite3 behaves.
So I decided to go with Docker. Docker is useful for running apps like these because it packages everything the app needs—code, libraries, and dependencies—into a container. This allows the app to be run across different environments (any user’s machine, irrespective of their device, OS, software etc.). Using Docker involved
- Creating a Docker file with the key commands for Python version, WORKDIR, EXPOSE of streamlit port and ENTRYPOINT
- Building the docker image locally
- Creating a new repository in Docker Hub
- Tagging the local image to the repository and pushing.
Observations
- The PDF documents should not have spaces in their titles. But this can be fixed with some string manipulation
- Sometimes the titles are not retrieved as expected but this is an issue with the PDF parsing using PyPDF more than the LLM itself.
Next steps
- The extraction from PDFs is most likely what is causing the inconsistency. My guess is that different PDFs may have different ways they were rendered - some were saved as PDFs from Word or Google docs, while others may have been formatted using LaTex or other designer software.
- So far I have only been able to extract the metadata of the papers into a tabular format. Now I want to explore if it would be possible to automatically format them into a set citation standard like APA, MLA, Chicago etc.
- This uses OpenAI API credits, so I want to find out if it can be done using only free resources – i.e. open-source LLMs and embeddings from Huggingface. (I did try fiddling around with Ollama but it was crashing my laptop quite bad so I switched to the simpler Huggingface models).
- I still need to deploy the Docker Image on a Cloud Platform so public users can access the dashboard.
- More to come…